LUBAC accelerates B-cell lymphomagenesis by conferring B cells resistance to genotoxic stress.
Blood 136(6): 684-697, 2020. DOI
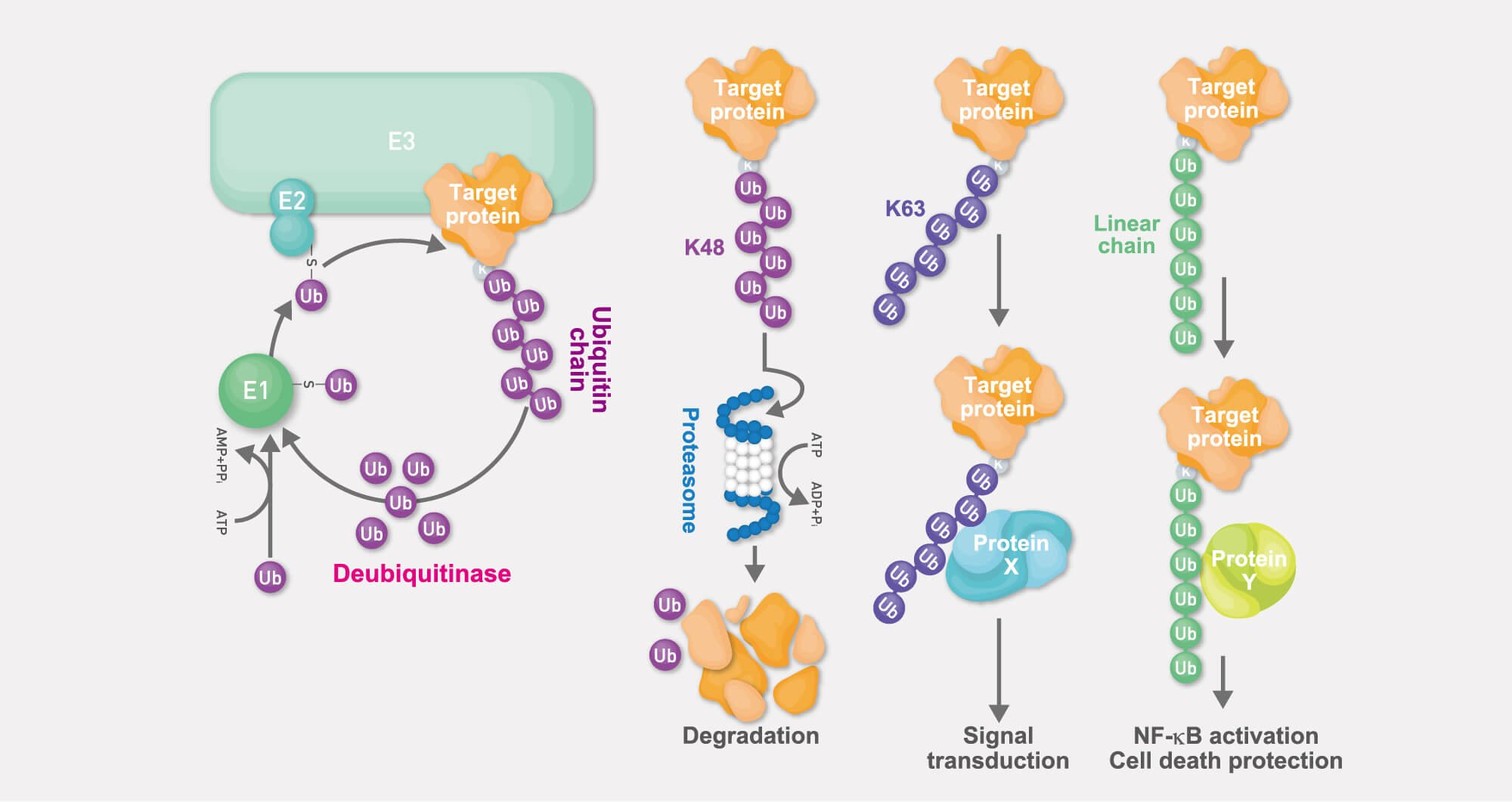
Ubiquitylation is one of the major post-translational modification systems, in which, via a cascade of reactions catalyzed by three types of enzymes (termed “E1”, “E2”, and “E3” enzymes), the E3 (ubiquitin ligase) target-specifically conjugates ubiquitin to substrate proteins.
Although originally the ubiquitin system had been identified as part of the cellular system of energy-dependent proteolysis (“ubiquitin-proteasome system”), the ubiquitin system is now well recognized as one of the most sophisticated reversible post-translational modification systems that regulates protein function in a wide variety of ways.
When compared to other post-translational modifications, the ubiquitin system shows several unique characteristics. They are:
For a long time it had been believed that ubiquitin chains are generated exclusively via one of ubiquitin’s seven lysine (Lys-linked) residues. In spite of this reigning paradigm, we discovered a brand-new type of ubiquitin chain: the linear ubiquitin chain. It is linked not via any of the Lys residues, but via the N-terminal methionine residue (Met1-linked). In addition to promoting pathophysiological analyses of these linear ubiquitin chains, we also discovered that abnormal generation of linear chains underlies various diseases including cancer and inflammatory diseases.
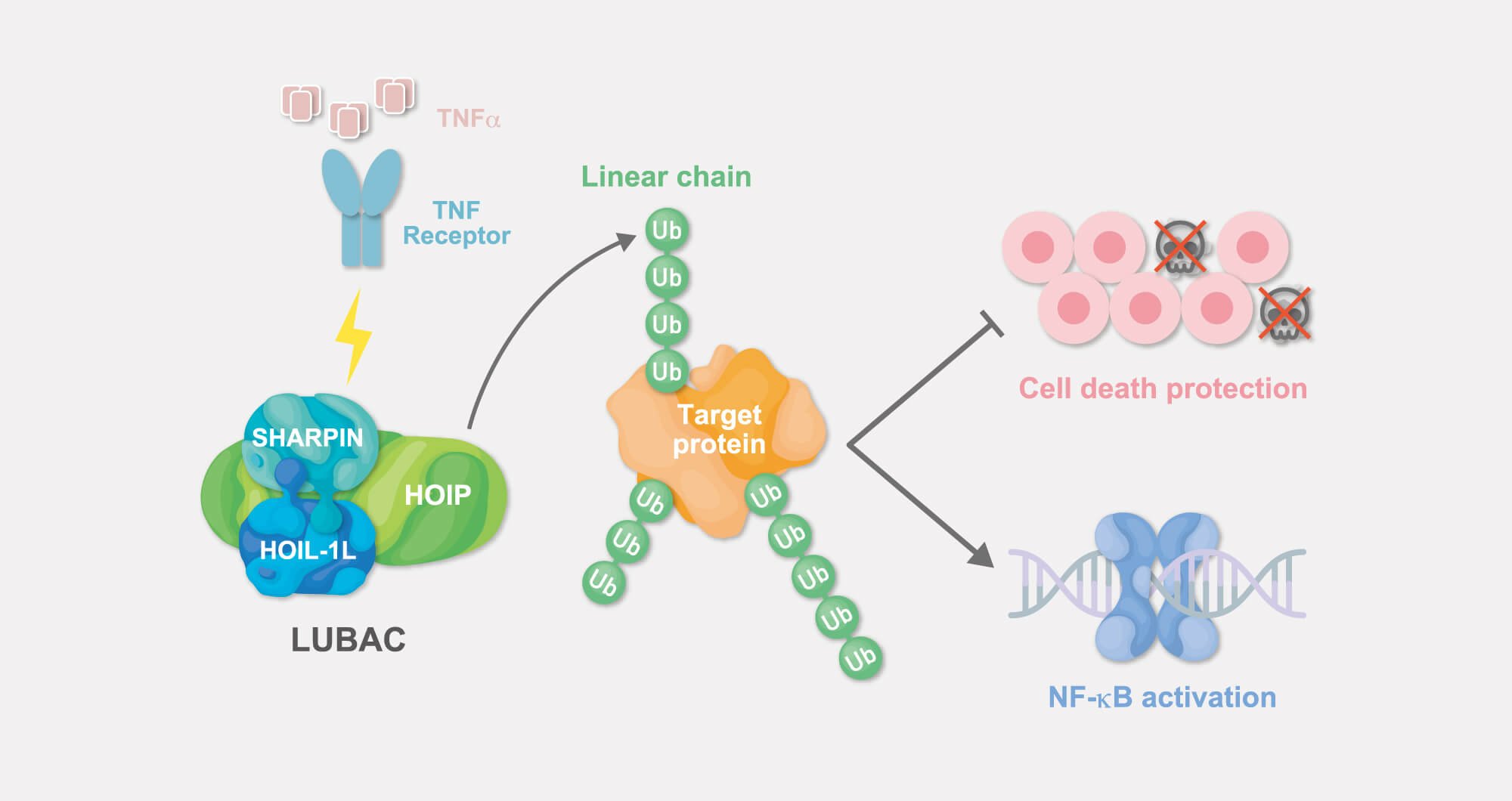
Functional analysis of linear ubiquitin chains
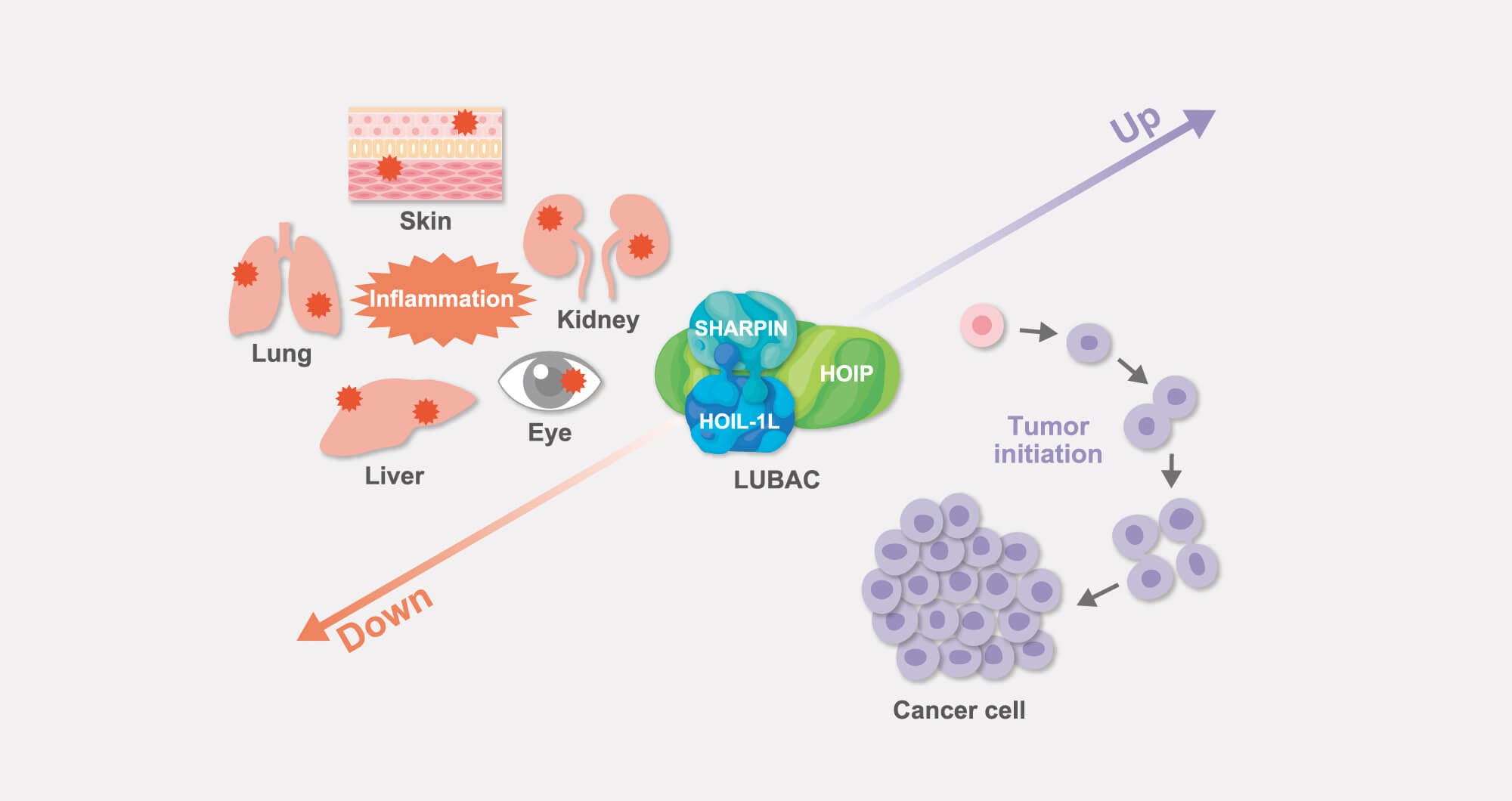
Role of linear ubiquitin chains in chronic inflammation and cancer
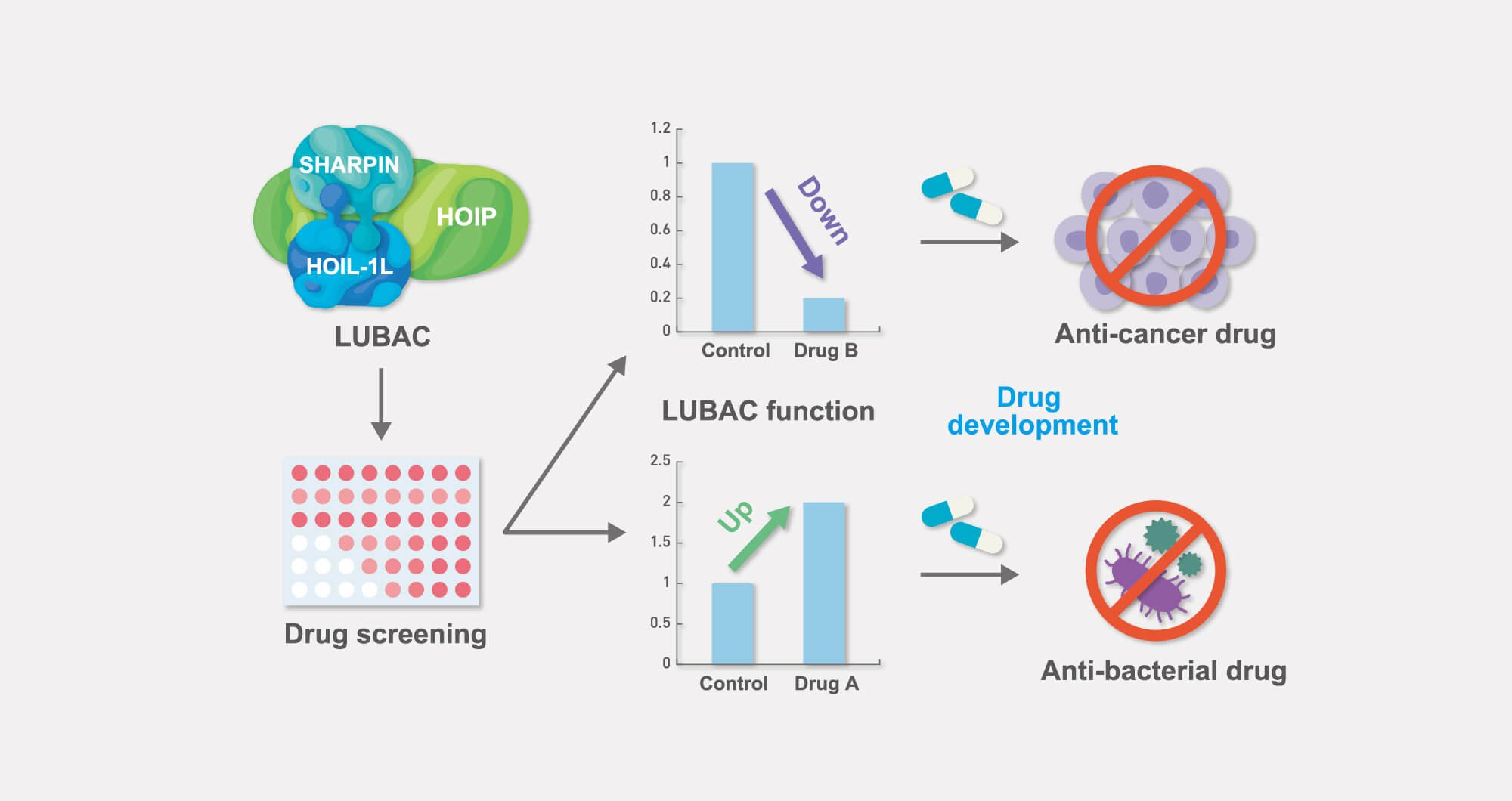
Screening to identify drugs that control the function of LUBAC
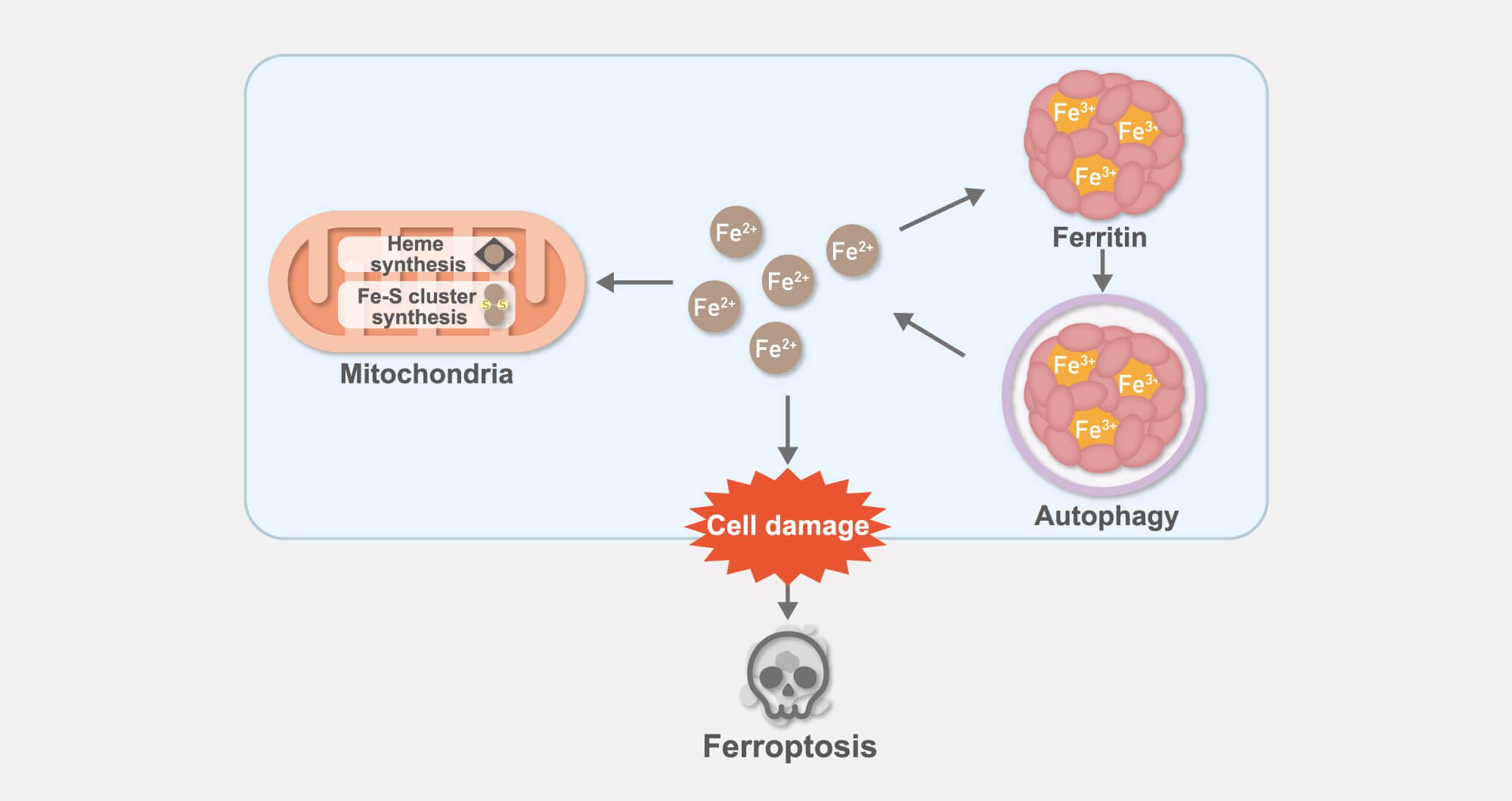
Analysis of intracellular iron dynamics and ferroptosis

So far, the LUBAC ubiquitin ligase complex is the only known (E3) ubiquitin ligase that generates linear ubiquitin chains.
Linear ubiquitin chains generated by LUBAC are involved in activation of the transcription factor NF-κB induced by various extracellular stimuli such as TNF-α, and also play a crucial role in suppression of programmed cell death.
By elucidating mechanisms underlying the regulation of LUBAC and identifying new proteins targets of LUBAC, we are currently exploring yet unidentified roles of linear ubiquitin chains and their contribution to the onset of disease.
Blood 136(6): 684-697, 2020. DOI
Nature Cell Biology 22(6): 663-673, 2020. DOI
Nature Commun. 10(1):3878, 2019. DOI
Cell Reports 23(4):1192-1204, 2018. DOI
J. Immunol. 200(10):3438-3449, 2018. DOI
Mol. Cell. Biol. 36:1569-1583, 2016. DOI
J. Immunol. 192:3793-3804, 2014.
Mol. Cell. Biol. 34:1322-1335, 2014.
Genes Cells. 19:254-272, 2014.
EMBO J. 32: 2463- 2476, 2013.
Proc. Natl. Acad. Sci. USA 108:20520-20525, 2011.
Nature 471:633-636, 2011.
Nature Cell Biology 11:123-132, 2009.
EMBO J. 25: 4877–4887, 2006.
Nature Cell Biology 5:336-340, 2003.

We have shown that, by regulating intracellular signal transduction systems, linear ubiquitylation plays crucial roles in both onset and maintenance (and even exacerbation) of the inflammatory environment in tissues in many immune and inflammatory diseases including cancer.
By utilizing organ-specific LUBAC-hyperactive and -deficient models, we are currently aiming at clarifying even more detailed physiological functions of the linear ubiquitin chain at the tissue level and at the level of individual animals. At the same time, we are analyzing new mechanisms that act in the pathogenesis of inflammatory diseases by applying various experimental approaches.
Blood 136(6): 684-697, 2020. DOI
Nature Commun. 10(1):3878, 2019. DOI
J. Immunol. 192:3793-3804, 2014.
Nature 471:633-636, 2011.
Nature 471:637-641, 2011.

In numerous previous studies, we and other groups have clearly demonstrated that both augmented and attenuated function of LUBAC can provoke various diseases.
We therefore aim at developing new therapeutics targeting LUBAC. Drugs that have the potency to inhibit LUBAC activity are likely to be suitable as anticancer drugs, whereas drugs that increase LUBAC activity are thought to be promising antibacterial drugs.
Based on our findings, we are now actively conducting drug screening assays to develop both LUBAC inhibitors and activators.
Blood 136(6): 684-697, 2020. DOI
Cell Reports 23(4):1192-1204, 2018. DOI
ACS Chem Biol. 10:675-681, 2015.
Cancer Discovery 4:480-493, 2014.

Iron is an essential nutrient for almost all living organisms. Conversely, it can also be toxic when present in excess. For this reason, intracellular iron metabolism must be tightly regulated.
We are studying the mechanism of iron homeostasis through the analysis of mitochondria, in which iron is integrated into iron-binding prosthetic groups such as the heme group, iron-sulfur clusters, and the iron-storage protein ferritin.
In addition, we are – from an iron-based perspective – extensively dissecting the molecular mechanisms underlying ferroptosis, which is an iron-dependent form of cell death that has been attracting tremendous attention in recent years.
Neurosci. Lett. 588:29-35, 2015.
Mol. Cell. Biol. 32:4998-5008, 2012.
Mol. Cell. Biol. 31:2040-2052, 2011.
Cell Metabolism 14: 339–351, 2011.
Mol. Biol. Cell 18:2980-2990, 2007.
Molecular Cell 19:171-181, 2005.
Proc. Natl. Acad. Sci. USA 95:4924-4928, 1998.
EMBO. J. 14:5350-5357, 1995.